This page was generated from: notebooks/nanopores/how_to_process_a_large_data_file.ipynb
[1]:
%load_ext autoreload
%autoreload 2
%config InlineBackend.rc = {'figure.figsize': (10,6)}
%matplotlib inline
Memory-map a large data set
This example demonstrates how to load and process a large data file (8 GB). This file is too large to load into memory at once (on most PCs), so memory mapping is used to only load the part of the data. A small volume and a single slice are extracted and saved for use in the other examples.
[2]:
import numpy as np
from nanomesh import Image
Load the data using mmap_mode='r'. This is a shallow interface to np.memmap that prevents the entire data set to be loaded into memory at once.
[3]:
data_name = 'G:\escience\hpgem\PG_EBAR_18072017_CH_6C_s15_10nm_rec_sa7_1024_1024_2048.vol'
vol = Image.load(data_name, mmap_mode='r')
vol
[3]:
Volume(shape=(2048, 1024, 1024), range=(-2034.4696044921875,1646.58935546875), dtype=float32)
[4]:
Image.load?
The show_slice method is still quite responsive to look at sections of the data. show_volume also works, but loads the entire volume into memory, which may make everything a bit slow and unresponsive 😅
[5]:
vol.show_slice(index=500)
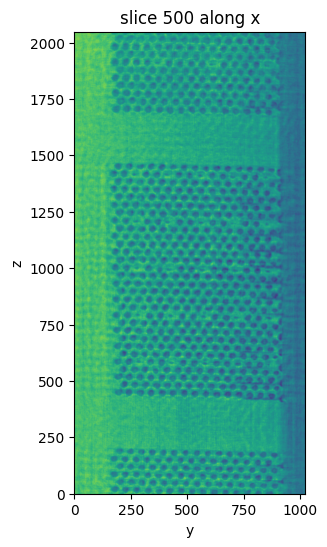
[5]:
<nanomesh.image._utils.SliceViewer at 0x2c62fec1070>
It’s easier to work with a section of the data. Note that a np.memmap object can be sliced like a normal numpy array, so we can extract a subvolume to work with:
[6]:
cropped = vol.select_subvolume(xs=(450, 550), ys=(150, 725), zs=(410, 1470))
cropped.show_slice()
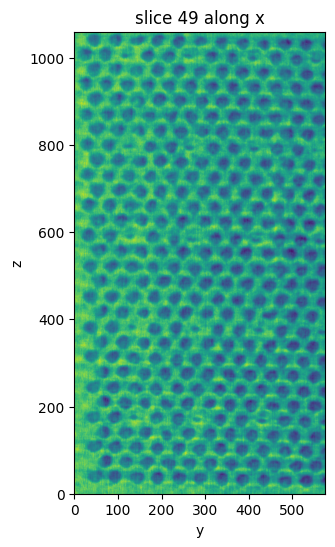
[6]:
<nanomesh.image._utils.SliceViewer at 0x2c83026dd90>
Display the cropped data.
[7]:
cropped.show()
Save the data to numpy binary format.
[8]:
cropped.save('slab_x450-550.npy')
Select a slice from the data and trim the edges for further analyses
[9]:
plane = vol.select_plane(x=500)
plane = plane.crop(left=150, right=850, top=655, bottom=845)
plane.show()
[9]:
<AxesSubplot:xlabel='x', ylabel='y'>
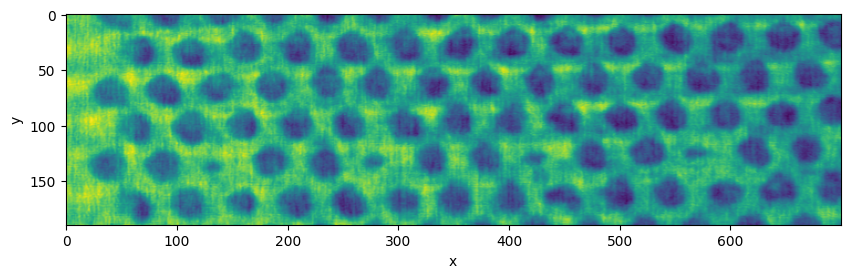
And save it…
[10]:
plane.save('nanopores_gradient.npy')