This page was generated from: notebooks/examples/how_to_do_3d_multi-domain_meshing_using_simulated_data.ipynb
[1]:
%load_ext autoreload
%autoreload 2
%config InlineBackend.rc = {'figure.figsize': (10,6)}
%matplotlib inline
Multi-domain mesh from simulated data
This notebook demonstrates an example using tetgen as the tetrahedralizer starting from a data volume.
Tetgen file formats: http://wias-berlin.de/software/tetgen/1.5/doc/manual/manual006.html#ff_poly
[2]:
import numpy as np
from nanomesh import Image
Generate data
The cell below generates a data volume with feature blobs. It consists of two domains, 1 for the bulk features, and 0 for the background.
If you want to use your own data, any numpy array can be passed to a `Image <https://nanomesh.readthedocs.io/en/latest/nanomesh.volume.html#nanomesh.volume.Volume>`__ object. Data stored as .npy can be loaded using Image.load().
[3]:
from nanomesh.data import binary_blobs3d
data = binary_blobs3d(seed=42)
vol = Image(data)
vol.show_slice()
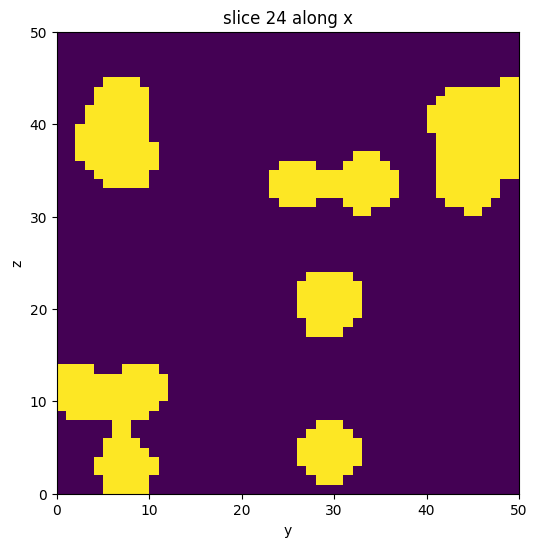
[3]:
<nanomesh.image._utils.SliceViewer at 0x21f972dd7c0>
Meshing
Mesh generation consists of two steps.
Generate the contour mesh
Generate the volume envelope
The contour mesh is the surface separating the domains in the data. In this case, the 1’s and 0’s, so the contour level does not need to be defined. The surface mesh is completed by wrapping the entire data volume in an envelope. This makes sure that the mesh (including any domain regions) is watertight, which is a requirement for generating a tetrahedral volume mesh.
[4]:
from nanomesh import Mesher
mesher = Mesher(vol)
mesher.generate_contour()
To plot the surface mesh:
(Use .plot_itk() for an interactive view)
[5]:
mesher.contour.plot_pyvista(jupyter_backend='static', show_edges=True)

Generating a tetrahedral volume mesh
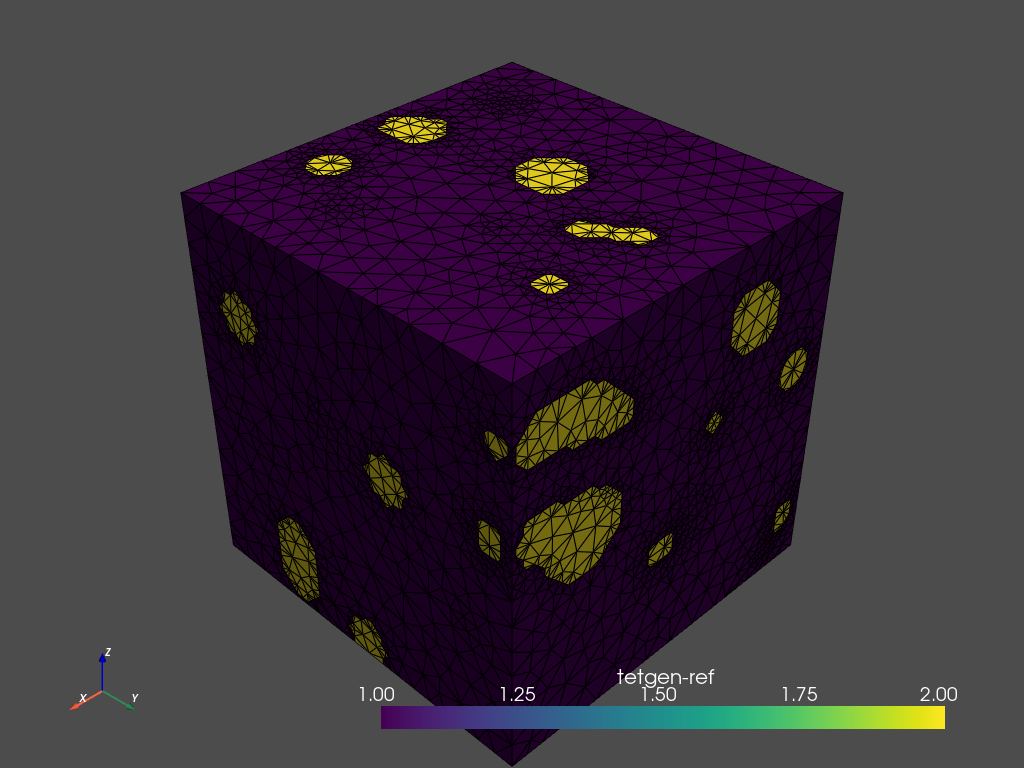
The volume mesh is generated using the .tetrahedralize method. This returns a tetrahedral mesh. Each domain, separated by the contour mesh defined above, is assigned a value.
The options used below:
-A: Assigns attributes to tetrahedra in different regions.-p: Tetrahedralizes a piecewise linear complex (PLC).-q: Refines mesh (to improve mesh quality).-a: Applies a maximum tetrahedron volume constraint.
Don’t make -a too small, or the algorithm will take a very long time to complete. If this parameter is left out, the triangles will keep growing without limit.
The region attributes are stored in the tetgenRef parameter.
Available options: http://wias-berlin.de/software/tetgen/1.5/doc/manual/manual005.html
[6]:
tetras = mesher.tetrahedralize(opts='-pAq -a10')
tetras.plot_pyvista(jupyter_backend='static',
show_edges=True) # Use .plot_itk() for an interactive view
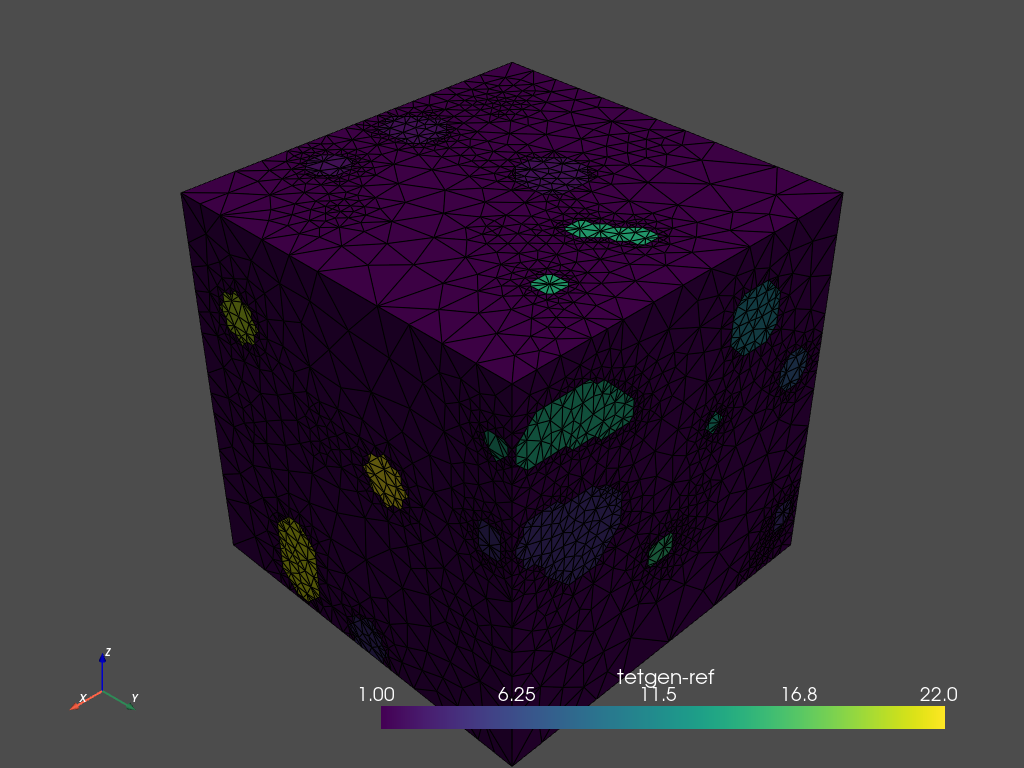
By default, the region attributes are assigned automatically by tetgen. Region markers assign attributes to tetrahedra in different regions. After tetrahedralization, the region markers will ‘flood’ the regions up to the defined boundaries. The elements of the resulting mesh are marked according to the region they belong to (tetras.cell_data['tetgenRef'].
It is possible to set your own attributes. The label corresponds to the attribute, and the value must be a point inside the region. The constraint can be used to set the maximum size of the triangles in combination with the -a parameter.
The next cell shows how you can update the region markers for the contour.
[7]:
for i, region_marker in enumerate(mesher.contour.region_markers):
if region_marker.name == 'background':
region_marker.constraint = 100
else:
region_marker.constraint = 1
region_marker.label = i + 1
region_marker.name = f'feature{i+1}'
mesh = mesher.tetrahedralize(opts='-pAq -a')
# Use .plot_itk() for an interactive view
mesh.plot_pyvista(jupyter_backend='static', show_edges=True)
[8]:
tetra_mesh = mesh.get('tetra')
for marker in mesher.contour.region_markers:
num = np.sum(tetra_mesh.cell_data['tetgen-ref'] == marker.label)
print(f'{num} tetrahedra with attribute `{marker}`')
112540 tetrahedra with attribute `RegionMarker(label=1, point=(11, 35, 24), name='background', constraint=100)`
20654 tetrahedra with attribute `RegionMarker(label=2, point=(19, 18, 40), name='feature2', constraint=1)`
576 tetrahedra with attribute `RegionMarker(label=3, point=(2, 15, 42), name='feature3', constraint=1)`
36897 tetrahedra with attribute `RegionMarker(label=4, point=(24, 14, 19), name='feature4', constraint=1)`
0 tetrahedra with attribute `RegionMarker(label=5, point=(5, 18, 16), name='feature5', constraint=1)`
1400 tetrahedra with attribute `RegionMarker(label=6, point=(4, 32, 27), name='feature6', constraint=1)`
577 tetrahedra with attribute `RegionMarker(label=7, point=(5, 47, 29), name='feature7', constraint=1)`
679 tetrahedra with attribute `RegionMarker(label=8, point=(7, 27, 43), name='feature8', constraint=1)`
671 tetrahedra with attribute `RegionMarker(label=9, point=(9, 41, 19), name='feature9', constraint=1)`
826 tetrahedra with attribute `RegionMarker(label=10, point=(13, 47, 40), name='feature10', constraint=1)`
683 tetrahedra with attribute `RegionMarker(label=11, point=(14, 38, 32), name='feature11', constraint=1)`
4648 tetrahedra with attribute `RegionMarker(label=12, point=(29, 4, 11), name='feature12', constraint=1)`
670 tetrahedra with attribute `RegionMarker(label=13, point=(19, 46, 29), name='feature13', constraint=1)`
7945 tetrahedra with attribute `RegionMarker(label=14, point=(35, 39, 43), name='feature14', constraint=1)`
1882 tetrahedra with attribute `RegionMarker(label=15, point=(25, 46, 16), name='feature15', constraint=1)`
644 tetrahedra with attribute `RegionMarker(label=16, point=(23, 38, 28), name='feature16', constraint=1)`
595 tetrahedra with attribute `RegionMarker(label=17, point=(37, 13, 2), name='feature17', constraint=1)`
451 tetrahedra with attribute `RegionMarker(label=18, point=(38, 2, 34), name='feature18', constraint=1)`
661 tetrahedra with attribute `RegionMarker(label=19, point=(40, 14, 45), name='feature19', constraint=1)`
478 tetrahedra with attribute `RegionMarker(label=20, point=(48, 8, 37), name='feature20', constraint=1)`
847 tetrahedra with attribute `RegionMarker(label=21, point=(47, 14, 7), name='feature21', constraint=1)`
489 tetrahedra with attribute `RegionMarker(label=22, point=(48, 31, 28), name='feature22', constraint=1)`